About us
RESULTS SUMMARY
The individual by individual results of current FTDNA testing are available at “DNA Results" on the menu to the left.
----------------------------------------------------------------------------------------------------------------------------------------------------------------------
TABLE OF CONTENTS
R1b "Tip of the Iceberg" Descendants Tree
R1b "Backbone" and Early Branching Timeline
R1b "Ancient DNA" Early Branching Tree
R1b Frequency % of the Total Male Population Map
R1b "Hall of Fame" Top Tested Surname Subclades
R1b All Subclades Project Detail Reports
R1b Summary Story by FTDNA with Ancient R1b DNA and Time Estimates
Major Subclade Projects within R1b
Other Sources of Information (includes link to R1b-YDNA discussion and resources group)
Analysis and Commentary on R1b Subclades
Please join R1b Y DNA project Facebook group for updates, questions and answers.
----------------------------------------------------------------------------------------------------------------------------------------------------------------------
R1b Tip of the Iceberg Descendant Tree
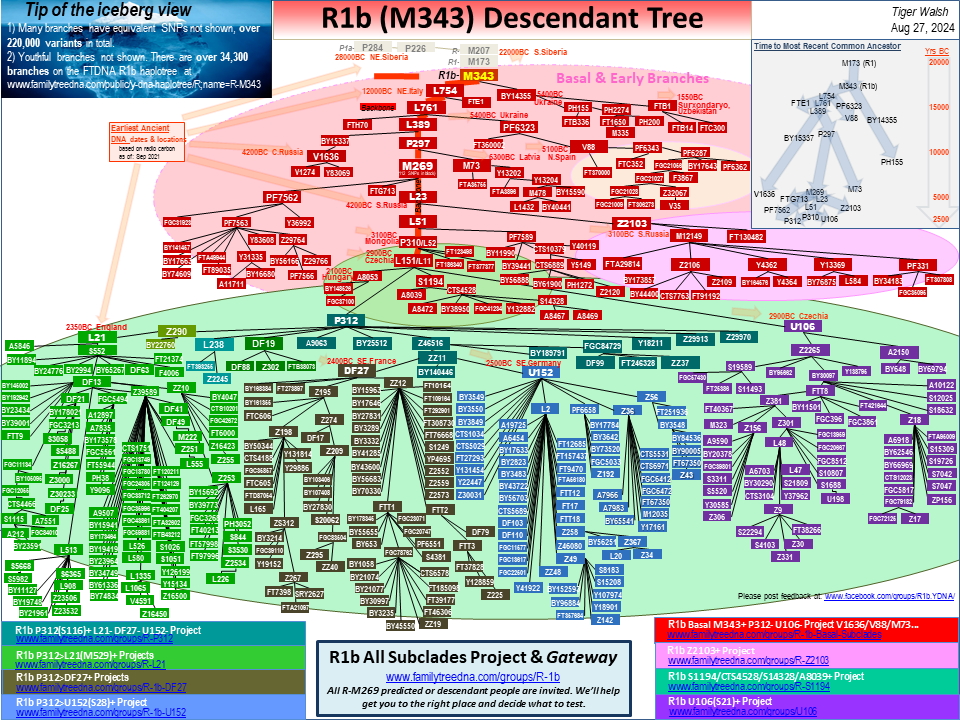
The Descendant Tree chart is just the tip of the iceberg. FTDNA's on-line Public Haplotree for R1b is detailed with tens of thousands of branches, reaching into genealogies. It is the largest and most current tree available. The projects listed form the R1b Project System.
Go to top
R1b Ancient Backbone and Early Branching Timeline
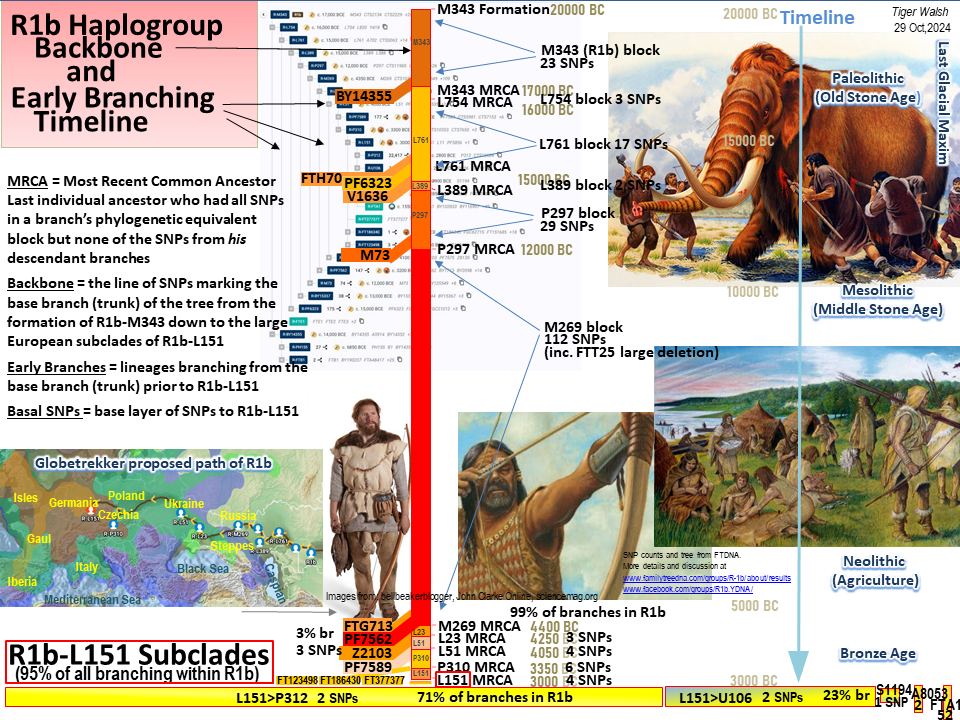
Go to top
R1b Ancient DNA Early Branching Tree
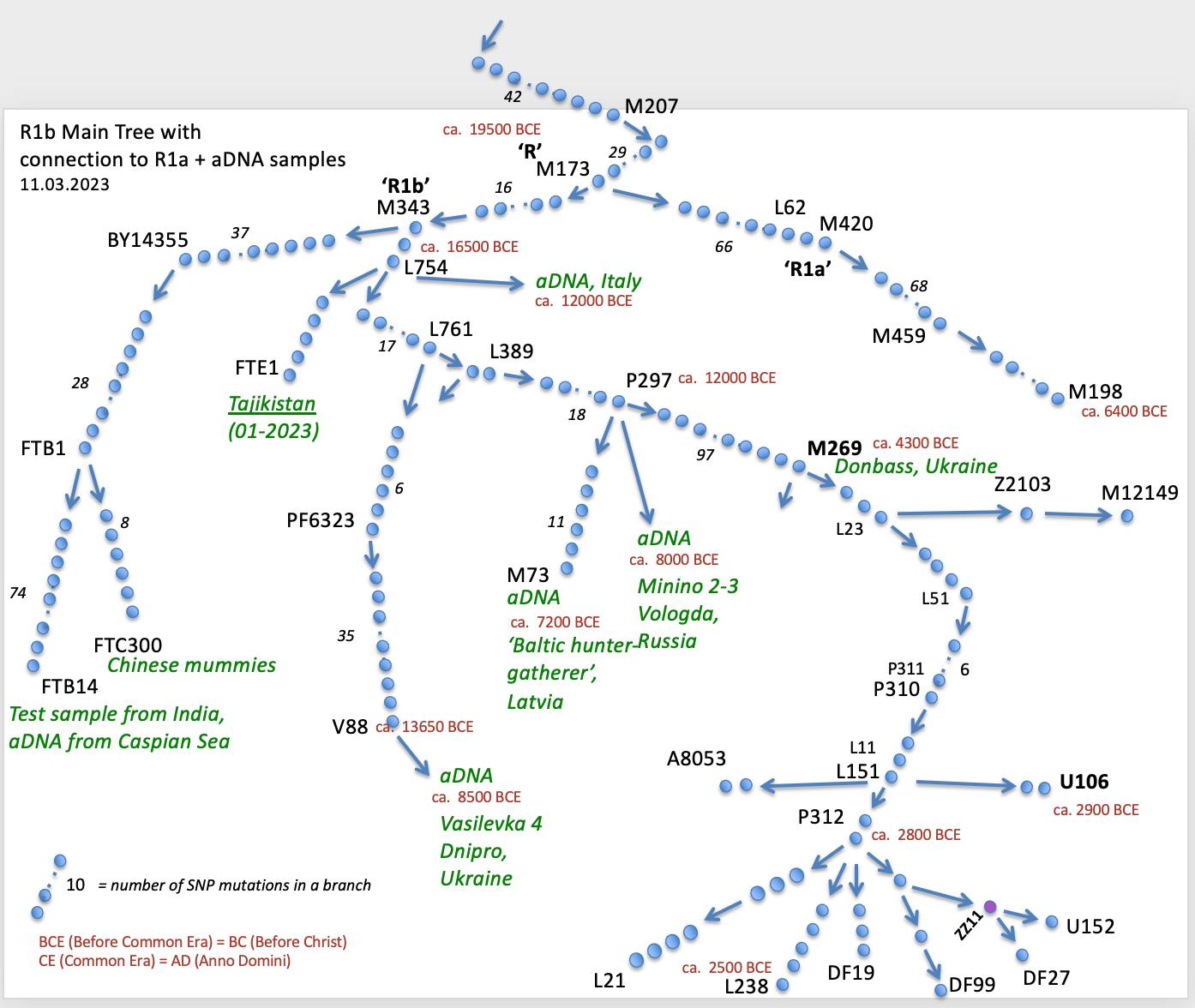
Go to top
R1b Frequency Percentages of the Total Population
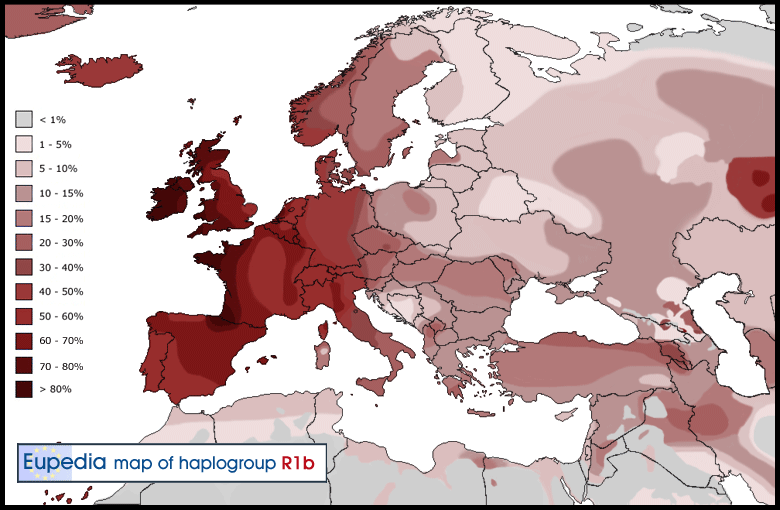
Go to top
R1b Top Tested Surname Subclades
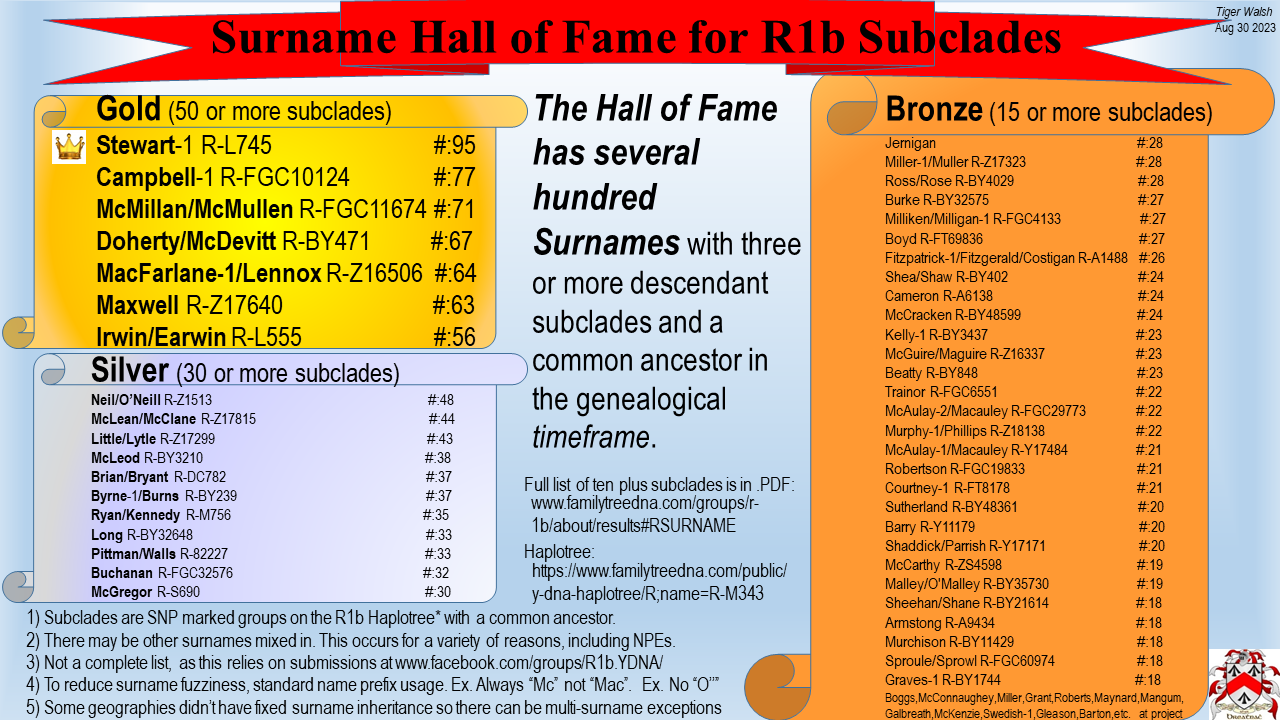
R1b Full Surname List (250+ names and haplotree URLs) in PDF format (download)
Go to top
Project Detail Reports
FTDNA's On-line Public Haplotree - An on-line version of the largest and most current Y DNA tree. It contains all the current branching, including equivalent SNPs, country, and surname reporting. Make sure you opt into sharing so that your branches will be counted correctly.
Gold Haplotypes file - An Excel based spreadsheet with every haplotype/kit in the project that have opted in to sharing and has either SNP tested to a major haplogroup level or has Y111 STRs. Earliest Known Ancestor, haplogroup and STR data is included. The focus is on Gold (Big Y700) and Gold Bridge (Y111) haplotypes.
You can review ancient clade mates beyond Big Y matching because the file is sorted in branch sequence number order according to the branching in FTDNA's haplotree.
You can look for matches beyond FTDNA's matching thresholds because a Y111 Genetic Distance calculator is included.
You can review summary STR statistics by name, country or subclade. The latest version is maintained on the R1b project Activity Feed on the Welcome post: https://www.familytreedna.com/groups/r-1b/activity-feed
Y Classic project chart - Detailed report of all Y STR values for every member in the projected sorted by project subgroup. Check this report for project administrator recommendations on STR and SNP testing. The report comes in multiple pages so as not to be slow, but you can view all of the pages at once if you enter 25000 into “page size.” If you are looking for a particular name or kit number you can do a “Ctrl-F” or “Find” from your internet browser. Many members use the Y Classic Chart to copy/paste (special) data from so they can do their own analyses. Please read the “Analysis and Commentary” (scroll down) for information on the subgroups. Here are the subgroupings used in the project.
100. M343+ M269- Subclades (R1b-M343,M73,V88,PH155)
110. M269+ L51- Z2103- Subclades (R1b-M269,PF7562)
120. Z2103+ Subclades (R1b-M343>L278/P25>L389>P297>M269>L23>Z2103)
130. L51+ P312- U106- S1194- Subclades (R1b-PF7589)
135. S1194 Subclades (R1b-M343>L278/P25>L389>P297>M269>L23>L51>P310>L151>S1194)
200. DF27+ Subclades (R1b>M269>L23>L51>L11>P312>ZZ11>DF27)
205. DF27+ Z209+ Subclades (R1b>M269>P312>DF27>Z209)
300. L21+ Subclades (R1b>M269>P312>L21 other)
305. L21+ DF21+ Subclades (R1b>M269>P312>L21>DF21)
310. L21+ DF41+ Subclades (R1b>M269>P312>L21>DF41)
315. L21+ DF63+ Subclades (R1b>M269>P312>L21>DF63)
320. L21+ FGC11134+ Subclades incl. CTS4466+ (R1b>M269>P312>L21>FGC11134)
325. L21+ FGC5494+ Subclades (R1b>M269>P312>L21>FGC5494)
330. L21+ L1335+ Subclades (R1b>M269>P312>L21>L1335)
335. L21+ L513+ Subclades (R1b>M269>P312>L21>L513)
340. L21+ M222+ Subclades (R1b>M269>P312>L21>M222)
345. L21+ S1026+ Subclades (R1b>M269>P312>L21>S1026)
350. L21+ S1051+ Subclades (R1b>M269>P312>L21>S1051)
355. L21+ Z251+ Subclades (R1b>M269>P312>L21>Z251)
360. L21+ Z253+ Subclades (R1b>M269>P312>L21>Z253)
365. L21+ Z255+ Subclades (R1b>M269>P312>L21>Z255)
400. U152+ Subclades (R1b>M269>L23>L51>L11>P312>U152)
500. P312+ L21- U152- DF27- Subclades (R1b>M269>L23>L51>L11>P312 & DF19,DF99,L238,BY22760)
600. U106+ Subclades (R1b>M269>L23>L51>L11>U106)
900. Unable to confirm Subclade; limited SNP testing [Consider Big Y700, or myFTDNA 111 matching]
910. Unable to confirm Subclade, limited STR/SNP testing [Consider Big Y700, or Y111 STR upgrades]
999c. R1b CONFIRMED (but not STR tested, Consider Big Y700, Need Y STR37,Y111)
X. Unsure of haplogroup [Consider Big Y700 or contact FTDNA] XX. M343- [Please withdraw from project]
Y Colorized project chart - Detailed report of all Y STR values for every member in the projected sorted by project subgroup. This report has statistics for each STR for each subgroup. This is includes the mode (most common), the minimum and the maximum values. Genetic genealogists often use the modal value as a proxy for the ancestral value for a subclade. This is not always true, but it is oftentimes very helpful to look for an a pattern of unusual values within a clade or subclade. This is called an STR signature and if you can identify a group that you fit into with a shared STR signature that can help your SNP testing be more efficient as the signature may indicate an underlying SNP marked subclade.
Y DNA SNP project report - Detailed report of all actual SNP test results for every member in the project. This report is very useful if you can determine a valid STR signature from the reports mentioned above. If you have other project members that you have close Genetic Distances (on your myFTDNA Y matches webpage) or you have members that match your STR signature you can search through this SNP report and see if your matches have tested for certain SNPs and see what their results are.
Ancestor project Map - Shows you the distribution of various project subgroups. You select which subgroup you want to view. You can use this to get an idea of where certain subgroups might be from. Be cautious, though, a point of origin is not necessarily the point with a high frequency of a particular subgroups. Many researchers think higher STR diversity and the presence of brother subclades is more indicative of origins.
Go to top
Major Subclade Projects
The discovery of SNPs that mark subclades of R1b has led to a number of major subclade projects. Depending on SNP results, members of the R1b project will be joined to one of these major projects. Data is integrated across these projects for analysis and understanding of R1b.
https://www.familytreedna.com/groups/R-1b-basal-subclades (R1b M343+ P312- U106- S1194- Subclades: V88, M73, M225, PF7562, PF7589, L23*, L51*, P310/L52*, L151/L11*. Basal = early branching)
https://www.familytreedna.com/groups/R-Z2103 (R1b-Z2103 and Subclades such as Z2109, L584)
https://www.familytreedna.com/groups/R-S1194 (R1b-S1194 and Subclades such as CTS4528/DF100, S14328, A8039)
https://www.familytreedna.com/groups/u106/about/results (R1b-U106 and Subclades such as L48, Z18, Z156, Z301, Z381)
https://www.familytreedna.com/groups/R-P312 (R1b-P312 and Subclades with focus on P312*, DF19, DF99, L238, ZZ37 or P312 unknown downstream)
https://www.familytreedna.com/groups/R-1b-U152 (R1b-U152 and Subclades such as L2, Z36, Z56, Z192)
https://www.familytreedna.com/groups/R-L21 (R1b-L21 and Subclades such as CTS4466, DF21, DF41, DF49xM222, M222, L1335, L513/DF1, Z251, Z253, Z255)
https://www.familytreedna.com/groups/R-1b-1c-7 (R1b-M222 and Subclades. M222 is the very large subclade of R1b-L21.)
https://www.familytreedna.com/groups/R1b-DF27 (R1b-DF27 and Subclades such as DF81, DF83, SRY2627, Z195, Z196, Z209, Z225, M153)
For help, please join the discussion group: https://www.facebook.com/groups/R1b.YDNA/
There are many other sub-projects but those listed above are the older haplogroups that are the next step down for new R1b/R1b1a2 people.
Go to top
Scientific Research Papers and Presentations
"The Beaker Phenomenon And The Genomic Transformation Of Northwest Europe" by Olalde, et al., 2017
http://biorxiv.org/content/early/2017/05/09/135962
"Genetic and the Peopling of Europe - Ancient and Modern Perspectives" by Hammer, et al., 2016
https://dl.dropboxusercontent.com/u/17907527/Genetics_and_Peopling_of_Europe_FTDNA_conf_by_Hammer_2016.pdf
"Ancient X chromosomes reveal contrasting sex bias in Neolithic and Bronze Age Eurasian migrations" by Goldberg, et al., 2016
http://www.pnas.org/content/early/2017/02/17/1616392114.abstract
"New clues to the evolutionary history of the main European paternal lineage M269: dissection of the Y-SNP S116 in Atlantic Europe and Iberia" by Valverde, et al., 2016
http://www.nature.com/ejhg/journal/v24/n3/pdf/ejhg2015114a.pdf
"Molecular Genealogy of a Mongol Queen’s Family and Her Possible Kinship with Genghis Khan" by Lkhagvasuren, et al., 2016
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0161622
"Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome" by Cassidy, et al., 2016
http://www.pnas.org/content/113/2/368.full.pdf
"The genetic Ice Age of Europe" by Fu, et al., 2016
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature17993.html
"R1b and the Peopling of Europe: An Ancient DNA Update" by Hammer, et al., 2015
http://www.slideshare.net/FamilyTreeDNA/r1b-and-the-people-of-europe-an-ancient-dna-update
"Large-scale recent expansion of European patrilineages shown by population resequencing" by Batini, et al., 2015
http://www.nature.com/ncomms/2015/150519/ncomms8152/full/ncomms8152.html
"Massive migration from the steppe was a source for Indo-European languages in Europe" by Haak, et al., 2015
http://www.nature.com/nature/journal/vaop/ncurrent/full/nature14317.html
"Origins of R-M269 Diversity in Europe" by Hammer, et al., 2013
https://gap.familytreedna.com/media/docs/2013/Hammer_M269_Diversity_in_Europe.pdf
"Discovery of Western European R1b1a2 Y Chromosome Variants in 1000 Genomes Project Data: An Online Community Approach" by Rocca, et al., 2012
http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0041634
"Emerging genetic patterns of the european neolithic: Perspectives from a late neolithic bell beaker burial site in Germany" byLee, et al., 2012
http://onlinelibrary.wiley.com/doi/10.1002/ajpa.22074/abstract
"The peopling of Europe and the cautionary tale of Y chromosome lineage R-M269" by Busby, et al., 2011
http://rspb.royalsocietypublishing.org/content/279/1730/884.abstract
"A Predominantly Neolithic Origin for European Paternal Lineages" by Balaresque, et al., 2010
http://www.plosbiology.org/article/info:doi%2F10.1371%2Fjournal.pbio.1000285
"A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe" by Myres, et al., 2010
http://www.nature.com/ejhg/journal/vaop/ncurrent/abs/ejhg2010146a.html
"Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152" by Cruciani, et al., 2010
http://www.ncbi.nlm.nih.gov/pubmed/20732840
>"Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages" by Cruciani, et al., 2010
http://www.nature.com/articles/ejhg2009231
"The Eurasian Heartland: A continental perspective on Y-chromosome diversity" by Wells, et al., 2001
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC56946/
"24,000 year old boy from Lake Baikal is scientific sensation" Siberian Times, 2015
http://siberiantimes.com/science/casestudy/news/24000-year-old-boy-from-lake-baikal-is-scientific-sensation/
"Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans" Raghavan, 2014
http://www.nature.com/nature/journal/v505/n7481/full/nature12736.html/
"Surfing during population expansions promotes genetic revolutions and structuration" by Excoffier and Ray, et al., 2008
http://www.cell.com/trends/ecology-evolution/abstract/S0169-5347%2808%2900167-5
"The Fate of Mutations Surfing on the Wave of a Range Expansion" by Klopfstein, et al., 2005
http://mbe.oxfordjournals.org/content/23/3/482.abstract
Go to top
Other Sources of Information
The FTDNA Activity Feed and the R1b-YDNA Yahoo Group are the primary communications vehicles for Q&A, daily conversations, project details and stats. The R1b Haplotypes spreadsheet in the Links section has statistics on mean, mode, median, standard deviation, allele distribution for each STR, by haplogroup and by geography as needed. More resources, links, analysis, diagrams and discussion are available at http://groups.yahoo.com/groups/R1b-YDNA/
Go to top
Analysis and Commentary on R1b Subclades
A comprehensive update on status of our knowledge on our haplogroup as of January 2011:
(Written by Tibor Fehér: Please note that the data below is my synthesis of what has been achieved by numerous professional and amateur genetic researchers, among whom there is no consensus on several aspects of the R1b history. Therefore, for everything written below, I take the responsibility when someone is in disagreement. Plus, all these should be treated as “the best what we can say know”, not as “unquestionable facts forever”. Outdated info removed.)
The Y haplogroup R1b (M343) appeared around 12,000 years ago in the Middle East, most likely somewhere in the Levant (today Syria, Lebanon, Israel, Jordan). These ancient forefathers were among those who developed farming in the area (but many other men of different haplogroups were also taking part.) We can distinguish three different subgroups with very different geographical distributions.
R1b-V88 (old R1b1c)
R1b-V88 chose to expand South-West into Africa and then along the Mediterranean coast to Spain and the Nile, reaching the Lake Chadareain Northern Cameroon and Northern Nigeria. This group is found rarely among Jews and other Mediterranean populations and is dominating the Chadic speakers of Sub-Saharan Africa
R1b-P25* (old R1b1*: P25+ V88- P297-)
R1b-P25* is ancestral to the bulk of downstream R1b clades. L278 has been found to be equivalent to P25 and is more stable. R1b-LP25* is rarely found in the Levant/Near East in its L389+ P297- form. Note that L389 is a recently discovered SNP that will help breakdown R1b-P25*. L278 appears to be an equivalent and more reliable marker for P25.
R1b-M73 (old R1b1a1)
R1b-M73 dispersed both into the West and East, reaching Spain and China, respectively. It is found rarely in the Mediterranean but several survivors went through a rapid demographic expansion in the Early Middle Ages in Inner Asia and expanded later with Turkic speaker nomads (Turkmens, Uzbek, Uyghur, Tatars, Cumans.)
R1b-M269 (old R1b1a2)
R1b-M269 is by far the most frequent subgroup today, more than half of Western European and (due to the colonization) American males belong to this line, including over 99% of the R1b project members. It is thought to have originated most likely in East Europe or West Asia around 8000 years ago(6500-8500 years with confidence). In its M269* (M269+ L23-) form, it is found rarely around the Mediterranean, Anatolia,the Balkans. We also have a Jewish cluster of R1b-M269*. “Ht35” is short for Haplotype 35, the original academic label for Eastern European and Asian R1b subgroups.
R1b-Z2103 (old L23* L23+ L51-)
R1b-L23* is relatively frequent in the Middle East (~10% in Turkey, Iran, Caucasus, Iraq, Syria, Jews and Albania) and dominant haplogroup of Armenians (20-30%) – it is thus called the “Armenian Modal Haplotype”. It is also found on the Balkans, where it has moved around 7000-8000 years ago, with early farmers. At the end of 2010, new SNPs were found in this group (L277, L405) which are not private and need further research to see how they split the L23* clade. People who are M269+ L23+ L51- P310- may test these SNPs with the chance of being positive.
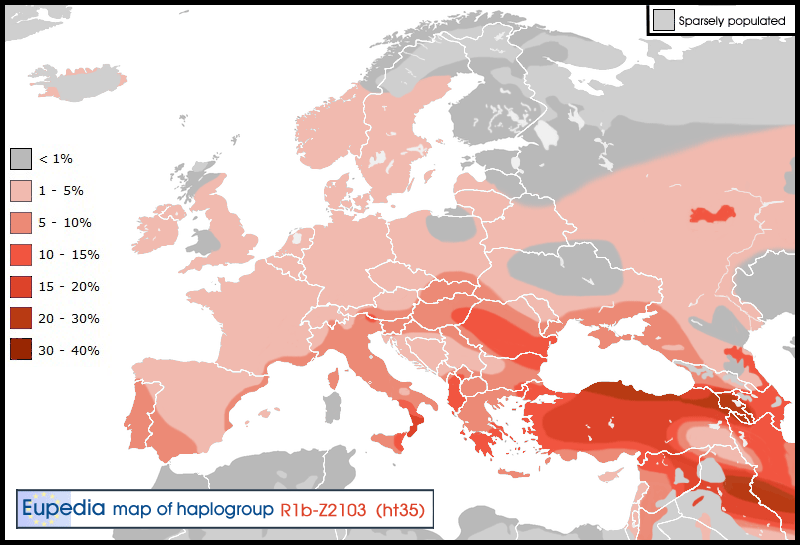
R1b-L51* (L51+ P311-)
R1b-L51* is an intermediary clade between the Eastern L23* and the Western L11+ (P310+) subgroups of R1b. It is found at extremely low frequencies from Iran to England, with relatively more occurrence in Central Europe. It is not yet clear if it arose in Europe or in Anatolia. People with the STRs of DYS426=13 and DYS393=13 have a good chance of longing here, but having such values does not mean you are surely L51* so deep SNP testing is always advisable.
R1b-P311 (P311) [S127, P310, L11 are just upstream of P311]
R1b-P311 is the parent of the “Atlantic Modal Haplotype” or"Western European R1b”. Around 50-60% of North Italian, Iberian,German,English and French (plus Colonial North and South American) men belong to this group, while its frequency exceeds 80% in more isolated areas like Ireland,Scotland, Wales, Bretagne and the Basque county. Its frequency is decreasing towards Scandinavia, East Central Europe and Southern Italy(20-30%), falling below 5% in Russia, the Balkans and the Middle East. It’s age is between 6500-5000 years BP or 4500-3000 BCE. It’s place of origin is likely Central Europe or the Lower Danube area, but there are claims for a more Western homeland (middle Rhine or Paris basin) as well. It is very rare in its P311*(U106- P312-) form, most people belong to one of the two big subgroups(U106 or P312). P311 and its subclades under went a rapid demographic expansion approx.5000-6000 years ago which resulted in the “invasion of Western Europe”, marginalizing the native inhabitants. It is disputed if this expansion correlated with the arrival of the Western Indo-European languages (Germanic, Celtic, Italic) – we should note that linkage between Y-DNA and languages is controversial, so I would leave this to what the reader thinks.
R1b-U106 (S21)
R1b-U106 is the Northeastern subgroup of R1b-L11. It is most common among Germanic speakers, with a frequency peak in the Netherlands (every third man is U106+) and decreasing frequency towards other Germanic-inhabited areas (15-25% in England, Germany, Austria, Switzerland and Denmark; 5-15% in Poland, France and Northern Italy; less than 5% elsewhere, including non-Danish Scandinavia). It is dated between 5500 and 4000 years ago or 3500-2000 BCE. One interesting feature of U106 is that it shows greater diversity in Poland and around the Baltic coast than in Germany and the Netherlands, so it may have appeared more to the East than its present distribution would suggest. It is yet debated if U106 people were the carriers of the proto-Germanic language or rather represent a Germanized Northern Celtic group of people or a Proto-IndoEuropean (PIE) folks later assimilated to Germans. U106 has several subgroups which are under research, including Z18, U198 (S29), L1 (null439),L48 (and its subgroups) and L257. None of these show a clear pattern which would make any connection to historical people possible.
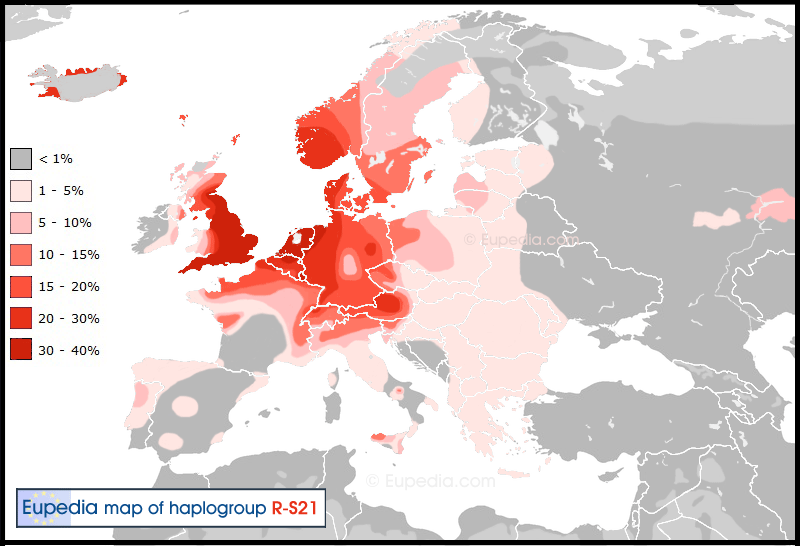
R1b-P312 (S116)
R1b-P312 is the Western subgroup of L11, the most common in Western Romance and Celtic-speaking areas. It has several subclades which have clear geographic pattern, the biggest being U152 and L21 (see later below). In its P312* (U152- L21-) form, it is most common in Iberia (35-50%) and France (20-35%), but also occurs frequently in Ireland, England and Switzerland(10-20%). It is less frequent (5-10%) in Northern Italy, Germany,the Netherlands and Scandinavia. Its small subgroup M153 is only found among Basques, while subgroup L176.2 consists of a Scandinavian (L165) and a Mediterranean (SRY2627/M167) branch. Its estimated age is similar to thatofU106, approximately 5500-4000 years before present (ybp) or 3500-2000BCE. Diversity points on France as the likely place of origin, and it is usually connected to the spread of Celto-Italic speaking populations. However, it may represent a more ancient folk (related to Basques) who only adopted Celtic languages and culture later.
R1b-P312>DF27
R1b-DF27 is a recently discovered subclade of R1b-P312 that has been found to encompass all of R1b-SRY2627 and R1b-M153. These have been considered Iberian Peninsular subclades but SRY2627 spreads as far north as Scandinavia and as far east as Germany. The old R1b North-South cluster is now also classified under R1b-DF27. (M.Walsh added DF27 section.)
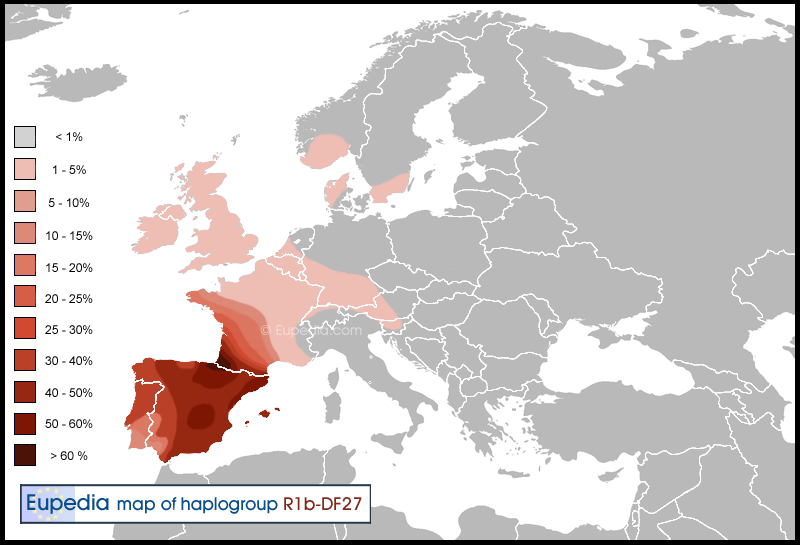
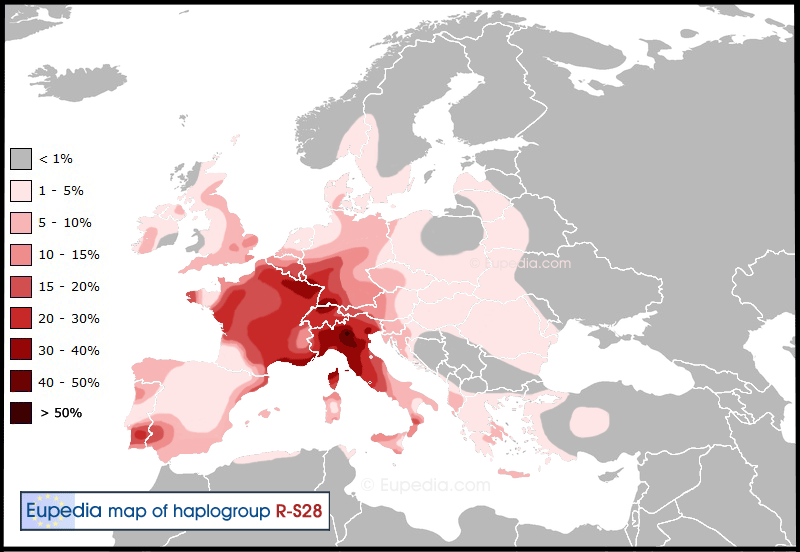
R1b-P312>U152 (S28)
R1b-U152 is the Alpine and Italian subclade of P312. It is most frequent in North Italy and Switzerland (20-40%) and in South Germany and France (10-20%) with decreasing frequencies towards neighboring areas (England, Low Countries, North Germany, Poland, Czech Republic, Hungary,South Italy, Iberia 5-10%). It also forms an isolated tribe among Bashkirs (Western Urals). Its diversity peak is in Germany and France, so up to our present knowledge, it more likely arose on the Northern slopes of the Alps,rather than the South of it. Its age is approximately 3500-5000 YBP or 3000-1500BCE. It is usually connected with the spread of the La Tene Iron Age archeological culture and historical Central European Celtic tribes, but it is nearly absent from present-day Celtic-speaking territories on the Atlantic coast. There are also theories connecting it to the spread of the Italic languages and later Roman colonization due to its Italian frequency, but these are not verified yet. Its main subgroups are the U152*, L2*, L20 and DYS492=14 groups, see the R1b-U152 project for more information.
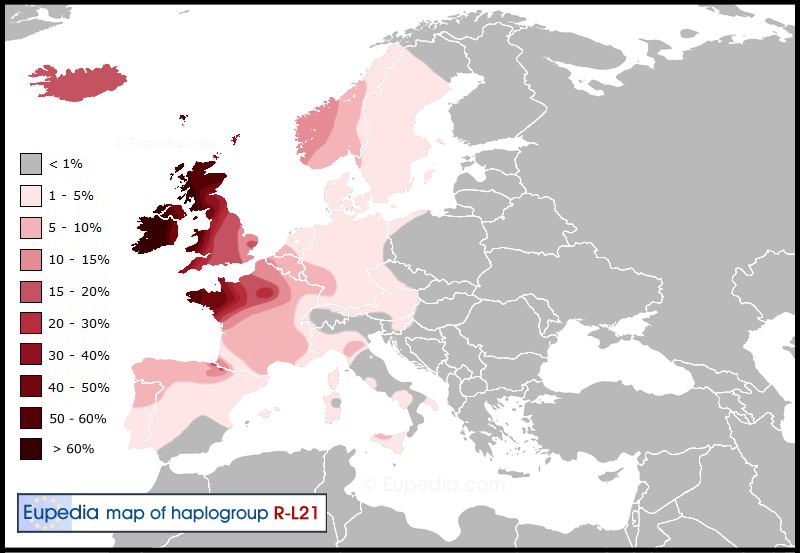
R1b-P312>L21 (S145, M529)
R1b-L21 is a predominantly British and Northwestern French subclade of P312 and is often referred to as a “Celtic marker”. It is most frequent in Ireland (50-90%), England (15-40%) and Bretagne (14%). L21 is also dominant among many North Americans due to their British Isles heritage. It also appears at reasonable frequencies in Norway and historically Celtic-inhabited areas of Iberia, France and Germany (2-10%), and more rarely in East Central Europe and Italy. Its estimated age is close to that of P312, approx.5500-4000years BP or 3500-2000 BCE. It is disputed when L21 reached the British Isles, some argue for a Bronze Age (Beaker) arrival, while others argue for a completely Celtic origin. L21 has several well-established subgroups like M222,L513, DF21, DF41, Z253 and Z255.
Other or predicted R1b
You must complete deep clade SNP testing to be placed into one of the subgroups listed above. The predicted only group of people are placed in “Y Classic Chart” #9 subgroups.